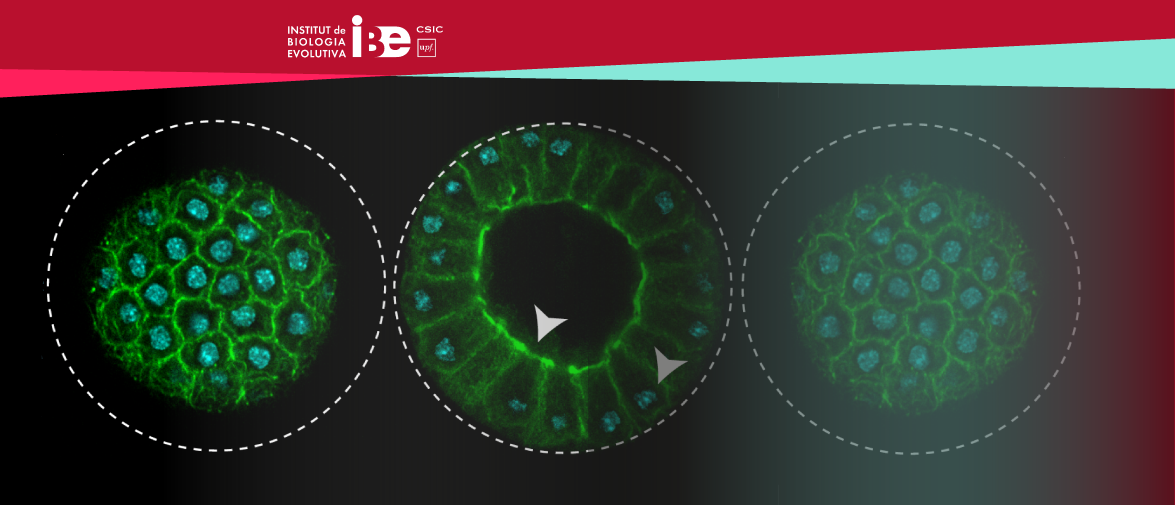
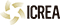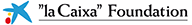El genoma de 240 espècies de mamífers revela què fa únic el genoma humà
La publicació especial de Science assenyala parts clau del genoma humà que s’han mantingut intactes durant milions d’anys.
L’Institut de Biologia Evolutiva (IBE, CSIC-UPF) i la Universitat de Pompeu Fabra han participat en la publicació especial de Science de la mà del Consorci Zoonomia. La publicació recull onze estudis internacionals i podria aportar nova informació sobre malalties i trets inusuals del genoma humà que es desconeixien fins ara.
Els estudis d’aquesta publicació aconsegueixen comparar els genomes de 240 espècies de mamífers, 4 vegades més que cap investigació anterior, aportant un gran recurs per a la comprensió i conservació de la biodiversitat genòmica. A més, fer disponibles els genomes de tantes espècies que difereixen en fenotips d’interès biomèdic (longevitat, taxes de neoplàsia, fertilitat, concentració de biometabòlits) permet l’estudi detallat de caràcters molt rellevants per la salut animal i humana.
Arcadi Navarro i Tomàs Marqués-Bonet, professors de recerca ICREA, investigadors de l’IBE (CSIC-UPF) i Catedràtics en Genètica de la Universitat de Pompeu Fabra, participen en el consorci internacional de Zoonomia, el qual constitueix el recurs comparatiu de genòmica de mamífers més gran del món. Mitjançant l’alineació del genoma de 240 espècies de mamífers, s’han identificat diferències en el seu ADN que poden desencadenar malalties o disfuncions greus en humans.
El projecte Zoonomia ofereix, de manera més detallada que en cap estudi anterior, un recurs que permet estudiar les regions reguladores de l’ADN, força desconegudes fins ara i que són difícils d’identificar amb genomes aïllats d’una sola espècie. D’aquesta manera, la investigació ha detectat i comparat regions altament conservades entre espècies de mamífers que podrien complir funcions clau, així com part de la base genètica dels trets característics d’alguns mamífers.

Les regions genòmiques més conservades entre els mamífers compleixen funcions vitals
Mitjançant la comparació de genomes de 240 espècies de mamífers, la investigació ha identificat dotzenes d’elements reguladors del genoma. Algunes d’aquestes regions genòmiques no han patit variacions en milions d’anys d’evolució, i podrien desvetllar funcions de vital importància per al correcte funcionament dels mamífers, donada la seva persistència. De fet, i gràcies a l’ús de tècniques de Machine Learning, l’estudi ha aconseguit vincular algunes d’aquestes regions conservades amb caràcters fonamentals, com ara la mida del cervell.
Per aquest motiu, l’estudi suggereix que petites variacions genètiques en aquestes regions d’ADN altament conservades tenen moltes probabilitats de causar malalties rares i comuns en els humans, com el càncer.
“Aquestes investigacions demostren, per tant, la utilitat d’utilitzar la conservació del genoma per l’estudi de malalties humanes, donat que es podria facilitar la detecció dels canvis genètics que augmenten o disminueixen el risc d’una malaltia en concret i que tots els membres d’una mateixa espècie comparteixen.” Assenyala Arcadi Navarro, que ha participat en la investigació de la mà del consorci Zoonomia.
Compartim un 10% de l’ADN amb la resta de mamífers
Segons aquests estudis, almenys un 10% del genoma humà es conserva simultàniament entre espècies de mamífers, entre les quals s’inclouen el gos, el ratolí, el ximpanzé i el ratpenat. Aquest percentatge es tradueix en més de 4.500 elements gairebé perfectament conservats en el genoma del 98% de les espècies estudiades.
La gran majoria d’aquests elements es troben fora de les regions d’ADN que codifiquen per proteïnes, el camp més estudiat de l’ADN i, en canvi, codifiquen per elements reguladors de la informació genètica, uns grans desconeguts per a la comunitat científica.
“Quan estudies el genoma aïllat d’una sola espècie pots identificar fàcilment les proteïnes, perquè són regions molt marcades amb un principi i un final.” Explica Arcadi Navarro. “Les regions reguladores, en canvi, només es poden detectar de manera sistemàtica i massiva mitjançant la comparació genòmica de diferents espècies i observant la seva evolució. Si es conserven és perquè tenen alguna utilitat, si desapareixen és perquè no suposaven un avantatge evolutiu o el seu entorn ha canviat.”
D’aquesta manera, les regions més conservades del genoma estan involucrades en el desenvolupament embrionari i la regulació de l’expressió d’ARN, mentre que les que més canvien tenen a veure amb la relació de l’animal amb el seu ambient, com ara la resposta immunitària, el desenvolupament de la pell, l’olfacte i el gust.
La hibernació o un gran olfacte són variacions genètiques amb un avantatge evolutiu
Els estudis comparatius també poden detectar aquelles mutacions que sí que han perdurat en el temps dins una espècie, per la qual cosa s'intueix que suposen un avantatge evolutiu.
La investigació assenyala regions del genoma associades a uns pocs trets excepcionals d'alguns mamífers, com ara un cervell extraordinàriament gran, un sentit de l'olfacte superior o l'habilitat d'hivernar durant l'hivern.
D'altra banda, també s'han detectat que aquelles espècies amb menys canvis a les regions conservades del genoma tenen un major risc d'extinció. Per aquest motiu, l'estudi conclou que amb només un genoma de referència per espècie es podrien identificar les espècies en perill. Encara es necessita més investigació al respecte, però aquesta pràctica podria suposar un avenç en els esforços per conservar la biodiversitat.
Aquests descobriments provenen de l'anàlisi de mostres d'ADN recol·lectades per més de 50 institucions diferents arreu del món, entre les quals s'hi inclouen mostres del zoològic de San Diego, que va proporcionar molts genomes d'aquestes espècies amenaçades o en perill d'extinció.
Article de referència: Matthew J. Christmas1 †, Irene M. Kaplow2,3†, et al. Evolutionary constraint and innovation across hundreds of placental mammals. Science. Vo.380, NO. 6643, Online April 28, 2023. DOI: 10.1126/science.abn3943